Mitochondrial DNA variation provides a tool for identifying
introduced provenances: a case study in Norway spruce
Christoph Sperisen1*,
1 Swiss Federal Institute for Forest,
Snow and Landscape Research,
2 National Board of Forestry, Vallgatan 8,
3 Present Address: University Children's Hospital, University
of Zurich, Division of Metabolic and Molecular Pediatrics,
*Corresponding author: Email: sperisen@wsl.ch
Introduction
Human impact on forests alters the genetic structure of tree species in several ways. An important example concerns the movement of seed. Plantings with non-local seed may eliminate local patterns of variation and may influence adjacent, native stands by pollen and seed migration. Because introduced stands are generally less well adapted to the site of introduction than native stands, introductions may affect adaptive variation of native stands. These effects are expected to be most important where intensive plantings were done with seed originating from regions that have environmental conditions that are very different from those at the site of planting.
Norway spruce (Picea abies (L.) Karst.) is a prime example of an intensively planted tree species. The species has been planted for several hundred years, and the introduction of foreign seed has been common since the beginning of the nineteenth century (Schmidt-Vogt 1977). Long distance seed movement has mainly occurred into northern Europe, where thousands of millions of trees have been planted from seed that originated from central and southeastern Europe (Laikre and Ryman 1996).
In recent years, the identification of native stands has become an important issue, mainly in the context of gene conservation. Native stands are generally considered to be valuable genetic resources, and their conservation is incorporated into many national conventions. Traditional methods, however, often do not allow to distinguish between non-native and native stands. Therefore, alternative methods are needed. In this study, we used a mitochondrial DNA marker for investigating the introduction of seed from central and southeastern Europe into northern Europe. Mitochondrial DNA is maternally inherited in Norway spruce and thus disperses only through seeds (Sperisen et al. 1999). The spatial distribution of mitochondrial DNA variation therefore allows direct inference on seed dispersal. The mitochondrial DNA fragment used in this study is the second intron of the nad1 gene. This intron contains six mutations which separate trees of northern Europe from those of central and southeastern Europe (Sperisen et al. 1998).
Materials and Methods
Eighty-seven samples were collected from clonal and plantation archives that were established from 16 old forests in Sweden located at sites where planting appeared unlikely. These gene conservation archives were established by the Swedish Forest Gene Conservation Programme. Two samples were collected from a stand planted with seedlings from seeds originating from southeastern Europe. 107 samples representing 18 provenances were collected from the International Union of Forest Research Organization (IUFRO) 1964/68 provenance trial for Norway spruce. This trial had an inventory character, and seed sources were sampled regardless of their native character (Krutzsch 1974). Our samples were collected in the Hungarian trial and were kindly provided by Éva Ujváriné and Béla Dudás. Total DNA was extracted as described (Ziegenhagen et al. 1993) and purified by using parts of the QIAamp blood kit (QIAGEN).
The second intron of the mitochondrial nad1 gene was amplified using a set of primers matching the nad1b and nad1c exons of angiosperms (Demesure et al. 1995). PCR-amplification was performed using a PTC-100 thermal cycler (MJ Research) in a total volume of 25 µl containing 1 x PCR buffer (Sigma), 1.6 mM MgCl2, 0.1 mM each of dATP, dCTP, dGTP and dTTP (Promega), 0.2 µM of each primer, 40 ng of template DNA, and 2.5 U of Taq DNA polymerase (Sigma) with the following profile: 3 min denaturation at 94°C and 41 cycles of 1 min denaturation at 94°C, 1 min annealing at 57°C, 2 min extension at 72°C followed by a final extension at 72°C for 8 min. The PCR products were digested with the restriction enzyme RsaI to screen for one of the six mutations which differentiate between Norway spruce of northern Europe versus Norway spruce of central and southeastern Europe. RsaI digestion reveals a 33 bp insertion/deletion sequence. The presence of this sequence is characteristic for Norway spruce of central and southeastern Europe and its absence is characteristic for Norway spruce of northern Europe.
Results and Discussion
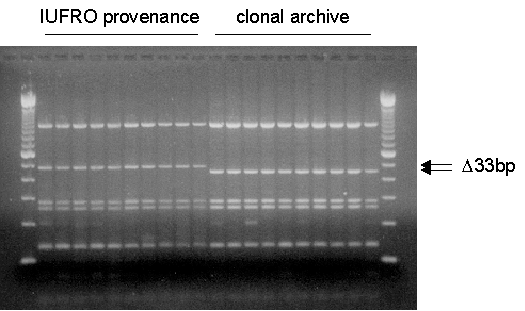
A total of 187 trees were screened for the presence/absence of the 33 bp sequence as revealed by restriction enzyme analysis (Figure 1).

Figure 1: Restriction fragment polymorphism of the second intron
of the mitochondrial nad1 gene of Norway spruce using the restriction
enzyme RsaI. Central and southeastern European trees contain a 33
bp sequence motif which is not present in northern European trees. Shown
are samples of a IUFRO provenance of southern Sweden and samples of a native
stand of southern Sweden which was used to establish a clonal archive.
Lanes 1 and 22: 100-bp ladder.
All trees sampled from gene conservation archives did not show the 33 bp sequence and thus conformed to the pattern we observed across northern Europe. The two trees of the stand planted with seedlings from seeds originating from southeastern Europe revealed the 33 bp sequence. The origin of this stand thus could be confirmed. Of the 18 provenances analysed from the Norway spruce provenance trial, 15 did not show the 33 bp sequence, while the two remaining provenances revealed the 33 bp sequence. These two provenances are located in southern Sweden (Figure 2) and most likely have been introduced from central or southeastern Europe. Natural immigration from central Europe to Sweden can be ruled out, because fossil pollen has been found neither in Denmark nor in the coastal area of southern Sweden.
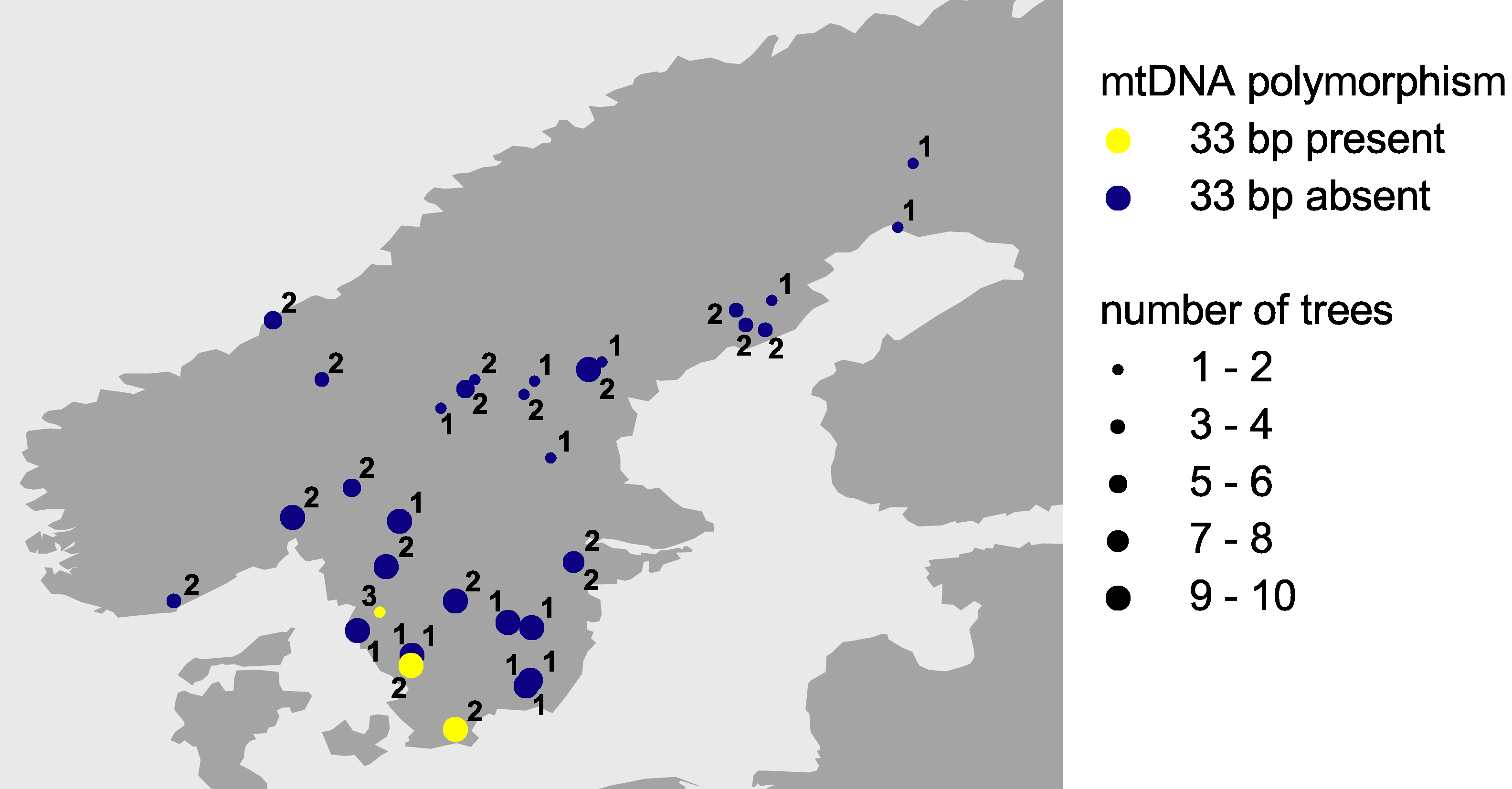
Figure 2: Polymorphism across northern Europe for the second
intron of the nad1 gene of Norway spruce. Yellow symbols represent
mitochondrial DNA containing the 33 bp sequence. Mitochondrial DNA lacking
this sequence is represented by blue symbols. Numbers give details on the
samples analysed: 1, stands from which gene conservation archives were
established; 2, IUFRO provenances; 3, stand planted with seedlings from
seeds originating from southeastern Europe.
Our results demonstrate the utility of mitochondrial DNA for the identification of long distance seed movements in Norway spruce. In this study, a single mutation was analysed, and assessment of more variation will be required in order to detect seed movement over shorter distances. The intron we analysed contains two short tandem repeats which are highly polymorphic (Sperisen et al. 1999). Their variation should prove useful for further subdividing the spatial pattern of the mutations which differentiate between Norway spruce of northern Europe versus central and southeastern Europe.
All the trees analysed from the gene conservation archives were of northern European origin. Several of these archives were established from forests in southern Sweden, where extensive planting of Norway spruce has occurred. Although old forests were selected at locations where planting appeared unlikely, one could not rule out that at least some trees were of central or southeastern European origin. Our results indicate that at least a large portion of the trees in the gene conservation archives are of northern European origin.
Our marker allows to distinguish unequivocally between trees of northern European origin and trees of central and southeastern European origin. The marker thus can be used to establish detailed geographic maps of native and non-native stands in regions where long distance seed movement has occurred. Such maps will be useful not only for developing management strategies for intensively managed forest areas but also for the conservation of genetic resources.
References
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology 4: 129-131.
Laikre L, Ryman N (1996) Effects on intraspecific biodiversity from harvesting and enhancing natural populations. Ambio 25: 504-509.
Krutzsch P (1974) The IUFRO 1964/68 provenance test with Norway spruce (Picea abies (L.) Karst.). Silvae Genetica 23: 1-3.
Schmidt-Vogt H (1977) Die Fichte. Band I. Verlag Paul Parey, Hamburg.
Sperisen C, Büchler U, Mátyás G (1998) Genetic variation of mitochondrial DNA reveals subdivision of Norway spruce (Picea abies (L.) Karst.). In Karp A, Isaac PG, Ingram DS (eds.). Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp. 413-417.
Sperisen C, Büchler U, Mátyás G, Anzidei M, Madaghiele A, Skrøppa T, Vendramin GG (1999) Polymorphic tandem repeats in the chloroplast and mitochondrial genomes of Norway spruce. In Skrøppa T, Paule L, Gömöry D (eds.). Genetics and Breeding of Norway spruce. Arbora Publishers, Zvolen, in press.
Ziegenhagen B, Guillemaut P, Scholz F (1993) A procedure for mini-preparations of genomic DNA from needles of silver fir (Abies alba Mill.). Plant Mol. Biol. Rep. 11: 117-121.
© Institut für Forstgenetik und Forstpflanzenzüchtung, Universität Göttingen, 1999