Julian P. Robinson, Stephen A. Harris*
Department of Plant Sciences, University of Oxford,
*Corresponding author: Email: stephen.harris@plant-sciences.oxford.ac.uk
Preamble.
Harris (1999a) reviewed the use of randomly amplified polymorphic DNA (RAPD) markers in systematic studies. RAPD techniques were embraced due to the relatively high levels of polymorphism they revealed and their low cost compared to other techniques, such as allozymes and restriction fragment length polymorphisms (RFLPs; Francisco-Ortega et al., 1993). Previously a lack of variation had been a considerable problem for many population studies. For example, in Red Pine (Pinus resinosa Ait.; Pinaceae) allozyme studies revealed no isozyme diversity (Fowler and Morris, 1977; Simon et al., 1986), but RAPD markers revealed diversity amongst red pines (De Verno and Mosseler, 1997). Two new marker methodologies appear to be supplanting RAPD analyses; Amplified Fragment Length Polymorphisms (AFLPs) and Simple Sequence Repeats (SSRs; microsatellites). This paper reviews these two techniques, the basis of the polymorphisms they identify; their advantages and disadvantages; their methods of analysis and their applicability to systematic studies.
Whilst the RAPD technique is fairly simple, both AFLP and SSR protocols are technically demanding (Karp et al., 1997). Both require competent users with experience in molecular biology techniques, although the skills required are falling with the emergence of kits for parts of the techniques (e.g. Perkin Elmer; www.perkin-elmer.com). The practical details for both techniques have been given in extensive practical reviews (Matthes et al., 1998; Morgante et al., 1998; Karp et al., 1997) and are both currently being experimented with to improve resolution, production of polymorphisms, cost and ease of use. A brief overview of the technique is provided, as it is essential that before using any technique the nature of the data that is generated should be understood.
Much has been written about the AFLP technique, most of it overwhelmingly positive. Much less has been written about microsatellites and phylogeny reconstruction, which is probably due to the expensive start-up costs and the supposed taxonomic limitations of SSRs. However, techniques must be critically evaluated, and the nature of the problem and the method used must be matched; research should be problem-solution, rather than technology, driven. Many researchers are aware of the limitations of methods and markers, but pragmatism often plays a major role in the choice of technique for a particular study.
AFLPs.
Vos et al. (1995) described the AFLP technique as being based on the detection of restriction fragments by PCR amplification and argued that 'the reliability of the RFLP technique is combined with the power of the PCR technique'. Other recommendations include those of Powell et al. (1996) who suggested that AFLPs 'provide high levels of resolution to allow delineation of complex genetic structures', whilst Winfield et al. (1998) saw AFLPs as 'reliable and informative multilocus probes'.
What are AFLPs and how are they produced?
AFLPs are fragments of DNA that have been amplified using directed primers from restriction digested genomic DNA (Matthes et al., 1998; Karp et al., 1997). Figure 1 explains the AFLP methodology:
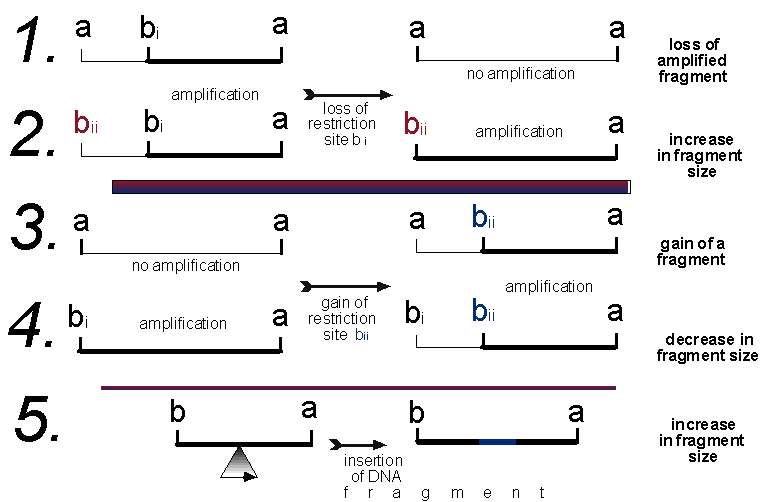
Figure 1. Five possible scenarios that will cause a change in
an AFLP profile. In scenarios 1 and 2 restriction site bi is lost, in 3
and 4 restriction site bii is gained, and in scenario 5 an insertion event
has taken place. Also applicable to scenario 5 are deletion and duplication
events, which will cause a decrease and increase in the fragment size respectively.
How AFLPs have been used.
Vos et al. (1995) were primarily interested in genome mapping using AFLP markers, i.e. construction of high density genetic maps of either genomes or genome fragments; 'it can bridge the gap between genetic and physical maps' (Vos et al. 1995). Since then many studies have applied this technique to mapping studies, e.g. Oryza (Zhu et al., 1998), Zea (Xu et al., 1999) and Solanum (Bradshaw et al., 1998). Xu et al. (1999) suggest that using AFLP is the most efficient way to generate a large number of markers that are linked to target genes. However, the high polymorphism revealed by AFLPs has also interested researchers from other fields. These fall into a few broadly defined categories: population genetics; phylogeny analysis; and cultivar/accession identification. It is the use of AFLPs in population genetics and phylogenetic analysis that are considered here.
At present the majority of population genetic uses of AFLPs are for "basic" diversity and genetic variation studies, as is often the case for new techniques. For example, Russell et al. (1999) investigated the genetic variation of Calycophyllum spruceanum (Rubiaceae), a fast growing pioneer tree of the Amazon Basin. Other studies have more specific goals such as investigations into introgression and hybridisation, e.g. Rieseberg et al. (1999) looked at introgression between cultivated sunflowers and a sympatric wild sunflower Helianthus petiolaris (Asteraceae) and Beismann et al. (1997) looked at the distribution of two Salix species and their hybrid. AFLP markers have also been used at the level of the individual, for use in paternity analyses and gene-flow investigations, e.g. Krauss & Peakall (1998) analysed paternity in natural populations of Persoonia mollis (Proteaceae), a long-lived fire-sensitive shrub from southern Australia.
Several studies have used AFLP markers in phylogenetic analyses. Heun et al. (1997) used a phylogenetic analysis of the allele frequency at different AFLP loci to suggest that Triticum monococcum subsp. boeticum (Poaceae) was the likely progenitor of cultivated einkorn wheat varieties. Others, including Aggarwal et al. (1999) who investigated the "phylogenetic relationships among Oryza species" using AFLP markers and Kardolus et al. (1998), who applied AFLPs to Solanum taxonomy, concluded that AFLPs were "an efficient and reliable technique for evolutionary studies". In the above papers the aims of the investigation were apparent from the title. However, other researchers use AFLPs to create dendrograms, but then suggest evolutionary hypotheses and correlate AFLP pattern similarity with phylogenetic closeness (e.g., Aggarwal et al., 1999; Mace et al., 1999).
A general feature of these investigations is that the species or genera have been investigated before using other markers; few AFLP studies have provided insights that were not available from other markers. For example, the genus Oryza has been intensively studied, and the results of Aggarwal et al. (1999) are generally consistent with Oryza taxonomy based on other lines of evidence, although Aggarwal et al. (1999) do not attempt to analyse either the O. sativa or O. officinalis complexes, both taxonomically unresolved areas. With a few exceptions (e.g. Russell et al., 1999; Muluvi et al., 1999), AFLPs have not been used to investigate the systematics of taxonomically complex or unknown groups, e.g. Eucalyptus, Carex, Solanum brevicaule sens. lat.
Advantages.
The major advantage of the AFLP technique is the large number of polymorphisms that the method generates. Its ability to differentiate individuals in a population makes the technique useful for paternity analyses (Krauss, 1999), gene-flow experiments, and also for Plant Variety registration (Law et al., 1998). Barker et al. (1999), investigating genetic diversity in Salix (Salicaceae), found 170 polymorphic bands with 20 RAPD primers, but 645 polymorphic bands with four AFLP primers. Nakajima et al. (1998) found that AFLP methods produced on average four times as many bands per reaction compared to RAPDs in their analysis of Daucus (Apiaceae) diversity. Similarly, Maughan et al. (1998) found that AFLPs produced more polymorphic loci per primer than RFLPs, SSRs or RAPDs in their study of Soybean (Glycine max and G. soja; Leguminosae) diversity.
Other advantageous features of the AFLP technique are: i) no sequence information is required; ii) the PCR technique is fast; and iii) a high multiplex ratio is possible (Rafalski et al., 1996).
The lack of sequence information needed by the AFLP method is similar to that of the RAPD technique. This is contrary to RFLPs and SSRs that need a high degree of characterisation of the target genome. This advantage is diminished as more taxa are examined, and as the database of characterised organisms grows and "universal primers" are discovered. For example, Taberlet et al.'s (1991) plastid trnL primers amplify a variable cpDNA region, approx. 1.75kb in length across a wide range of genera and families [e.g. Ginkgo biloba (Ginkgoaceae), Rosa canina (Rosaceae) and Phalaris arundinaceae (Poaceae)]. In the case of SSRs the range of conservation is less, generally being restricted to other species within the same genus (e.g. Steinkellner et al., 1997) or tribe (e.g. Dayanandan et al., 1997).
Since the AFLP technique is PCR-based it can provide high throughput; Krauss and Peakall (1998) suggest that, after the initial screening period, up to 100 individuals for 100 polymorphic loci per week could be analysed. This makes it ideal for large-scale population studies. Additionally, dried material can be used for the analyses (Russell et al., 1998), since the method is DNA-based. This enables analysis of species that would be difficult to sample ex situ (Harris and Robinson, 1994).
The multiplex ratio is the number of different genetic loci that may be simultaneously analysed per experiment (Rafalski et al., 1996). Since AFLP markers are distributed across the genome they have a high multiplex ratio, i.e. each band is assumed to come from a different area of the plant genome. In contrast, SSRs have a large number of alleles per locus, and have a multiplex ratio of 1 (Harris, 1999b). A high multiplex ratio is considered desirable, since it suggests that the whole genome is being sampled rather than one segment of it.
Problems
The problems associated with AFLPs can be divided into three types: practical; data; and analysis. Many of these problems are not unique to AFLP methodologies, but apply to most molecular marker systems. An ideal marker would have: sufficient variation for the problem under study; be reliable; and be simple to generate and interpret. Unfortunately, an ideal marker does not exist for use in all studies; rather a technique or techniques will be suited to a of range investigations (Karp et al., 1997; Harris, 1999b).
Practical Problems.
Many of the practical problems associated with AFLPs, unlike those of RAPDs, can be overcome, although users must be proficient in many practical skills.
Cost. Assigning a cost to process is always a tricky issue, since cost depends on your viewpoint and the amount of resources available. For example, the RAPD technique is fairly cheap, but in terms of data quality, the money may be wasted. There are, however, several expensive components in an AFLP analysis. The biotinylated probes and streptavadin magnetic beads are expensive, though increasingly a pre-selection amplification step is used (Vos et al., 1995). The primers and adapters may also be expensive, the cost of which varies depending on whether custom or `off-the-shelf' primers are used. However, the detection system, either g33P or fluorescent labels, will be fairly expensive (Huang and Sun, 1999). Since there are many variables, assessment of costs is difficult, Karp et al. (1997) estimate ECU 1.4 per assay. This is a generous estimate, more realistic costs are ECU 9.0 for the initial preselection (Krauss and Peakall, 1998), whilst we would estimate that initial start-up costs to be approximately ECU 12.0 per individual, falling to approximately ECU 5.0-7.5 per primer combination once the screening of primer combinations has finished and four different reactions are analysed in a single gel lane.
Restriction Enzymes and Primers. The choice of either restriction enzyme or primer can affect the number of AFLP polymorphisms detected. For example, more polymorphisms are detected in barley with the combination of restriction enzymes PstI/MseI than with EcoRI/MseI (Ridout and Donini, 1999).
The choice of primer may also have a large influence on the amount and quality of variation uncovered. For example, Hartl and Seefelder (1998) found in their analyses of hop (Humulus lupulus, Cannabaceae) cultivar diversity that [adapter]+2 primers produced too many bands to score or separate on a polyacrylamide gel, [adapter]+4 primers did not "exploit the resolution capability" of AFLPs, but [adapter]+3 primers produced a large number of defined bands. Hartl and Seefelder (1998) evaluated 60 primer combinations; only eight of these combinations provided reliable banding patterns. Lercerteau and Szmidt (1999) analysed 64 [adapter]+3 primer combinations (eight for each restriction enzyme adapter), of which 12 generated easily readable patterns, 17 could possibly have been used and 35 were excluded because of their complexity. Additionally, Lercerteau and Szmidt (1999) investigated the addition of a nucleotide to an [adapter]+3 primer; the [adapter]+4 primer reduced the number of bands found by 50%. Interestingly, the [adapter]+4 primer also amplified several bands that were not amplified with the [adapter]+3 primer. In Kardolus et al.'s (1998) study, one [adapter]+3 combination gave a "dense fingerprint" so that reliable scoring was not possible, but the other three [adapter]+3 combinations were scored. Thus, the choice of primer may influence the number of bands amplified and the level of polymorphisms found, which in turn is linked to the taxonomic level of the investigation. In general, for studies at the species level it would appear that [adapter]+2 primers amplify too many bands and [adapter]+4 primers do not reveal enough polymorphism, whilst some [adapter]+3 primers do not give patterns that are easily analysed. In general, the plant genome is AT-rich, so the use of AT-poor primers may reduce banding pattern complexity (Qi and Lindhout, 1997).
The above examples illustrate that not all primer combinations produce either reliable or sufficient data. If no preliminary screening of primer combinations is performed, then the choice of primers is essentially random and there is a risk that the primers will give insufficient data for analysis. Often the cost of the AFLP technique is such that only a limited number of primer combinations can be analysed and it may be tempting to analyse all of these combinations, even though some of them may be too "complex".
Reproducibility. AFLPs are generally acclaimed for their reproducibility, which sets the technique apart from RAPDs. Jones et al. (1998) tested the reproducibility of AFLPs throughout a network of European laboratories, and by rigorously controlling all the variables they were able to show that it is possible to reproduce AFLP banding patterns across a range of laboratories. Whether this level of rigor could be maintained in more test studies cannot be determined. Another aspect of reproducibility is reaction consistency, i.e. whether an accession and primer combination always gives the same results. Winfield et al. (1998) in an analysis of genetic diversity of Black Poplar (Populus nigra subsp. betulifolia, Salicaceae), ran duplicate samples for five trees (it was not indicated whether these were complete re-runs or re-amplifications); three duplicates returned exactly the same banding patterns, the other two were 98.9% and 97.6% similar. The number of different bands this equates to, and whether these differences were band gains or losses, were not documented. Krauss and Peakall (1998) encountered a "rare disappearing fragment", in which an initially polymorphic band was scored but subsequent analysis with a new DNA extraction and repeated AFLP amplification failed to reproduce this band. Krauss and Peakall (1998) suggest that this could be due to a partial digestion of the template genomic DNA, poor amplification of this fragment during PCR or DNA contamination. Furthermore, Donini et al. (1997) showed that tissue ontogenesis may influence AFLP patterns, due to the occurrence of organ-specific methylated restriction sites. Partial digestion appears to be the most common source of artefactual polymorphism in AFLP analyses (Lin and Kuo, 1995), and can be caused by a number of factors including "dirty" DNA or methylated DNA (Dowling et al., 1997).
The scoring of AFLP fragments, like RAPDs, is open to a certain amount of "interpretation". Travis et al. (1996) in an influential investigation of variation in the endangered plant Astragalus cremnophylax var. cremnophylax (Leguminosae) scored all "monomorphic and polymorphic fragments discernible in at least 95% of the individuals by eye". Escaravage et al. (1998), using fluorescent detection, rather than radioactivity, considered only "high intensity peaks" in their analysis of clonal diversity in a Rhododendron population. These chromatograms identified a mean of 50, 50 and 35, detectable peaks for each of the three primer combinations used, of which 25 (50%), 25 (50%) and 19 (54%) were analysed respectively. Angiolillo et al. (1999) found 288 polymorphic AFLP bands, in their diversity study of Olea yet only scored 121 (42%) bands that were described as unambiguous (Angiolillo et al., 1999); presumably the others were ambiguous. Aggarwal et al. (1999) only included "distinct, reproducible, well-resolved" fragments in their study of Oryza. None of the above studies, or any studies we reviewed defined what constituted a "distinct, reproducible, well-resolved" band.
Gift and Stevens (1997) investigated how different researchers delimited morphological characters in the genus Kalmia (Ericaceae). The authors asked 49 individuals to break the variation displayed in 10 characters (e.g. sepal width) into a series of discrete characters, and no two individuals scored the set of characters in the same way (Gift and Stevens, 1997). Gift and Stevens (1997) concluded that states in the data set were delimited in various ways by researchers, and the way that data was presented influenced the assignment of character-states. They also added that "expert knowledge" appeared to be of "dubious" value in delimiting states. Gift and Stevens' (1997) paper alludes to the problems of defining a "distinct, reproducible, well-resolved" band. No similar study, to the best of our knowledge, has taken place using a DNA fingerprinting technique (in the broadest sense of this term). Just as Gift and Stevens (1997) found it difficult for individuals to agree on character states, it may be expected that it would be difficult for a group of researchers to agree on what constituted a distinct, reproducible or well resolved band. It is anticipated the criteria for definition will be flexible according to the time and money allocated to a study. Krauss and Peakall (1998) recognised this and suggested that computer detection (as part of the fluorescent method of detection) of fragments is more efficient and accurate than scoring bands from autoradiographs.
The claims for AFLP reproducibility are well-founded, but there are several concerns: i) some studies have found different banding patterns when samples are re-run (e.g. Krauss and Peakall, 1998; Winfield et al. 1998); ii) the scoring of the AFLP bands and their inclusion in a data matrix is not explicit, which suggests that different individuals will score AFLP patterns differently; and iii) the method of genomic DNA preparation may affect banding patterns, e.g. partial digestion due to either poor DNA quality or insufficient (or faulty) restriction enzymes (Lin and Kuo, 1995).
Data problems.
Dominance. Like RAPD markers, AFLP markers are thought to be dominant, with polymorphisms detected as either band presence or absence. Dominant markers are not as efficient as co-dominant markers for population genetics studies (Lewis & Snow, 1992; Lynch and Milligan, 1994). Lynch and Milligan (1994) estimate that 2-10 times more individuals need to be sampled per locus for dominant markers compared to co-dominant markers. Krauss and Peakall (1998) suggest that this disadvantage may be overcome because of the large number of polymorphisms generated; over 100 polymorphisms per lane are possible. Maughan et al. (1996) in an AFLP study in Glycine also investigated the inheritance of AFLP markers. They examined six loci for one primer combination in 61 F2 plants. Five loci segregated in a dominant manner, whilst one locus appeared to be inherited in a co-dominant fashion. One of the causes of co-dominance was thought to be due to the presence of SSRs in the amplified fragments (Maughan et al., 1996), which has implications for the independence of AFLP markers. Therefore it is possible that an AFLP gel may have a mixture of dominant and co-dominant markers present on it, and without appropriate segregation analyses it will be impossible to determine marker segregation.
It has been suggested that it should be possible to identify heterozygotes from the intensity of the bands or peaks on AFLP gels (Castiglioni et al., 1999). That is, a heterozygote for a marker will have a band half as dense as the homozygote for the dominant allele, and is based on the idea that there will be twice as many markers for the homozygote dominant compared to the heterozygote. However, Vos et al. (1995) in their description of the AFLP technique state that the AFLP procedure is insensitive to template DNA concentration; similar band intensities were seen in a range of template concentrations (25ng-25pg; Vos et al., 1995).
Homology. Homology is perhaps the greatest problem in AFLP analysis. It is often assumed that co-migrating bands are homologous, though there is no a priori reason to accept this. Mace et al. (1999) suggest that the "mutual occurrence of several bands strengthens the likelihood of the pair-wise homology of all of them", although this does not appear a substantive argument for AFLP band homology. Furthermore, a particular sized band may consist of bands from different regions of the genome, and there is no way to assess the homology of missing bands, two different mutations could lead their absence. Rieseberg (1996), in a RAPD study showed that 91% of the co-migrating bands were homologous, though of these homologous bands ~13% were paralogous rather than orthologous. Kardolus et al. (1998) argue that the chance that two co-migrating AFLP fragments do not represent identical alleles of one locus is small, which they believe is due to the highly selective amplification and sharp resolution of polyacrylamide gel electrophoresis. Rouppe van der Voort et al. (1997), in a mapping study of Solanum (Solanaceae), sequenced 20 out of 117 putatively homologous AFLP markers, of which 19 (95%) were nearly identical. Given that 5% of scored bands may be non-homologous, the major issue is what the effects would be if this value were repeated throughout AFLP studies.
Lamboy (1994a, 1994b) considered this in detail for RAPD analyses, much of which is applicable to the problems encountered with AFLP analyses, and Bremer (1991) dealt with this in a discussion of RFLPs. Lamboy (1994a) suggested that artefacts, such as non-homologous products, would significantly bias genetic distance estimates between taxa. An additional consequence of using non-homologous (and non-independent) characters is the artificial increase in homoplasy, which may obscure the phylogenetic relationships of the taxa under investigation (Bremer, 1991).
Rieseberg (1996) suggested that RAPD homology was a function of taxonomic distance, i.e. the more closely the compared accessions are, the greater the probability that a shared co-migrating band is homologous. This may extend to the analysis of AFLPs, since Lerceteau and Szmidt (1999) found AFLPs to reflect the classical taxonomy of Pinus at lower taxonomic levels, but at higher taxonomic levels the data sets were incongruent; AFLP data placed Pinus sylvestris and P. merkusii closer to Picea abies than to Pinus gerardiana. Thus the issue of the taxonomic level at which AFLPs can be used is raised. This is not an simple question to answer, as levels of genetic similarity are not uniform throughout the plant kingdom, although a conservative view would be that above the species level the use of AFLPs to produce classifications and phylogenies is unwarranted.
Mutation rate. The level of polymorphism that different markers reveal is important. If the marker reveals too little variation, then it may not be possible to discriminate taxa. Unfortunately, if the variation found is too high then the relationships between the taxa tend to be obscured (Stuessy, 1990). This is caused by two factors: i) with high levels of variation the levels of similarity between two taxa are low, and both character and distance measures and tree reconstruction programmes are increasingly inaccurate at predicting relationships; and ii) if levels of variation are high then the probability of assigning correct homology is reduced. For example, the loss (or gain) of a band may be caused by the same event, so the taxa are not related by that event. The success of AFLPs is mostly due to the high levels of variation it reveals, which suggests that comparisons between distantly related taxa/accessions would produce inaccurate information about their relationships.
Scoring. Mutations are scored as presence or absence of a particular band, and from these observations a binary data matrix is built. At its simplest the differences between patterns are either the presence or absence of a restriction site. This may translate directly in to the presence or absence of a band on the AFLP gel, or to a change in size of the AFLP band (Figure 2). That is, three basic profile changes may occur, gain or loss of a band and change in the size of a band. Not all of these changes may be equally likely and will occur at different frequencies, although no one has investigated the frequencies of such events for AFLP data.

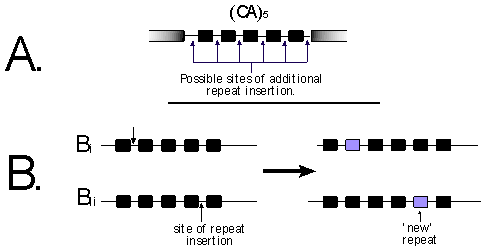
Figure 2. A. The black boxes are the CA nucleotides within
the amplified microsatellite. In this example there are five repeat units
(i.e. (CA)5), and the possible sites a "slippage" could
insert another repeat are indicated.
B.
The (CA)5 allele has gained another repeat, the
two illustrations indicate how the resulting (CA)6 alleles are
not homologous.
The loss of a restriction site is most likely caused by a point mutation in the restriction enzyme recognition sequence, causing the sequence not to be recognised by the enzyme and therefore not cut. Likewise the gain of a site is caused by a point mutation changing a potential site into a recognisable site. The probabilities of these two events are unequal (DeBry and Slade, 1985), the loss of a restriction site being much more likely than a gain. Site loss or gain may also be produced by an insertion, deletion or duplication event. Of the five scenarios predicted in Figure 1, only 1 and 3 will give "simple" changes to the data matrix, scenario 1 will be the loss of a band and 3 will be the gain of a band. Scenarios 2, 4 and 5 will cause two changes to the data matrix. It is likely scenario 2 will be scored as the loss of the smaller fragment (a b i) and the gain of a larger fragment (a b ii), vice versa for scenario 4. Furthermore, insertion, deletion or duplication events, causing a change in the size of a band, may be misinterpreted as a gain or loss of a restriction site/band. For example, an insertion event (scenario 5) may add a segment to a restriction fragment increasing its size. This would probably be scored as the loss of the smaller fragment and the gain of the larger fragment. Thus some of the AFLP data will be non-independent, which violates the assumption of independence amongst characters for phylogenetic analyses (Swofford et al., 1997).
Thus, unless the reasons for all the changes between all the taxa are investigated, which may be impossible, several changes are scored twice in the data matrix. The importance of this depends on the frequencies of the five scenarios; if 2, 4, or 5 occur at very low frequencies then the amount of disruption may be minimal. The mutation in scenarios 2, 4, and 5 that caused the change in the AFLP profile will be entered twice in the data matrix. Bremer (1991) suggests that these "overscoring" will be randomly distributed and so should not systematically bias the results, although she notes that weakly supported groups may be affected. Harris (1999a) indicated that clustering diagrams based on RAPD data often have long terminal branches and short internode distances, which also happens in AFLP analyses (e.g. Travis et al., 1996; Kardolus et al. 1998, Aggarwal et al. 1999). The non-independence of these scenarios may artificially increase the length of internodes, and suggest erroneous relationships. Swofford et al. (1997) were also unconvinced by Bremer's (1991) argument, and suggested that just because something is done inappropriately enough times there is no guarantee it will "work out in the end".
The ploidy level of the taxa under investigation may affect the amount of variation observed. Kardolus et al. (1998) in their investigation in Solanum recorded the mean number of polymorphic bands at each ploidy level. For diploids this was on average 112 polymorphic bands, for tetraploids 142 bands and for hexaploids 159 bands, although the differences between ploidy levels were not statistically tested. The extra bands, if correlated with polyploidy, introduce complications into their scoring in the data matrix. If these bands are only present in the polyploids then they cannot be scored as missing (0) in the diploid (and the tetraploid if they are hexaploid bands) but have to scored as undetermined (?). The absence or presence of bands in the diploids cannot be determined because the fragment that produces the band is not present in the diploid. This introduces difficulties when analysing diploids and polyploids in the same analysis.
Miscellaneous data problems. Barker et al. (1999) in their analysis of Salix species used five combinations of selective primers. Of these, four primers gave similar clustering patterns, a fifth combination suggested different patterns. Barker et al. (1999) ignored the final combination in their combined analysis, and speculated that the "discrepancy" related to skew in the distribution of the markers and illustrated the importance of testing several primer combinations. However, discordant data sets may give significant insights into a problem. There are possibly two explanations for the "discrepancy" observed by Barker et al. (1999), both of which illustrate issues pertinent to this discussion. In the dendrograms for the four primer combinations (Barker et al., 1999, their Fig. 3) the internode distances are small, especially those indicating basal similarities. It is possible that the fifth combination of primers produced a data set with problems such as non-independence and non-homologous fragments, which would introduce bias into the measurement of similarity.
The other second explanation raises another issue concerning the genomic origin of the fragments. Mapping studies have illustrated that most AFLP bands are distributed randomly across the genome (e.g. Zhu et al. 1998), though these studies group all their primer combinations together. It would be interesting to know if individual primer combinations showed a less random distribution, since some AFLPs appear to be non-randomly distributed (Rouppe van der Voort, 1998). The fifth primer combination in Barker et al.'s (1999) study may have had a different genetic history to that of the other primer combinations.
Data analysis.
Once AFLP profiles have been converted into a data matrix, then they can be analysed in one of three ways; similarity, frequency and character measures. Similarity or frequency measures convert the binary data matrix into a series of distance measures between taxa. The third method of analysis uses the data as characters in an analysis. The choice between similarity and frequency measures depends primarily on the number of accessions analysed and the aims of the investigation. When the number of accessions is small (<50) and the analysis is primarily focussed on variation between individuals, similarity measures tend to be used (e.g. Beismann et al., 1997; Escaravage et al., 1998). For those studies using larger numbers of accessions and with an emphasis on the variation between "populations", frequency measures are generally used (e.g. Perera et al., 1998; Muluvi et al., 1999). Character-based analyses are an unusual method of analysing AFLP data, except in those studies with an explicit phylogenetic hypothesis (e.g. Kardolus et al., 1998).
Similarity measures. Similarity measures used for AFLP data are: i) the simple matching coefficient (SMC; Sneath and Sokal, 1973), which measures the proportion of shared band presence and absences between two AFLP profiles; ii) Jaccard's coefficient (Jaccard, 1908), which measures the proportion of shared bands; or iii) the Nei and Li coefficient (NL; Nei and Li, 1979), which measures the probability that a band being amplified in one sample being amplified in another sample. NL also has a biological perspective, the coefficient is an estimate of the proportion of shared bands shared by two samples because they where inherited from a common ancestor (Harris, 1999a).
Lamboy (1994a) suggested that for RAPD data two major groups of error may be identified, false positives (a product is present but should be absent) and false negatives (a product that is absent but should be present). Both groups of error are certainly applicable to AFLP data, although at a lower level because of the stringency of the AFLP PCR reaction. The other sources of error are non-homology between co-migrating fragments, which could be classified as false positive, failure of complete digestion of genomic DNA and could be scored as either or both false positives or false negatives. In addition to these, there is the question of the non-independence of bands on an AFLP gel, which are neither false positives nor negatives.
All these sources of error introduce bias into the similarity estimates. Lamboy (1994b) calculated the bias for a range of possible scenarios varying the number of bands, the number of shared bands and the percentages of false negatives and positives. If it is assumed that the only source of error is that of non-homologous bands at a rate of 5% (Rouppe van der Voort et al., 1997) then the bias values calculated by Lamboy (1994b) range from 0.5% to 40% depending on the number of bands detected and the percentage of these that are shared. If Lamboy's data is applicable to AFLPs, then errors can have the potential to give inaccurate similarity measures, and consequently a large effect on the clustering of taxa. It is particularly a problem when the distance between internodes is small; as a small change here may alter the way groups cluster.
Frequency measures. Like RAPDs and allozymes, AFLPs are used for the assessment of "genetic diversity" within and between species, cultivars and populations. From the frequency of AFLP products, the levels and patterns of diversity are calculated. The markers are usually treated as independent and diversities are is calculated using: i) similarity measures (e.g. Russell et al., 1998); ii) Shannon's measure (e.g. Maughan et al., 1996; Zhu et al., 1998); or iii) analysis of molecular variance (AMOVA, Excoffier et al., 1992; e.g. Travis et al., 1996).
The sources of error are essentially the same for frequency measures, as with similarity measures, but with the additional complication of the dominance of the AFLP bands. With dominant loci the variance at a single loci is increased and introduces bias into the frequency measure (Lynch and Milligan, 1994), although there are methods to overcome this problem (Lynch and Milligan, 1994; Zhivotovsky, 1999). Clark and Lanigan (1993) enumerated nine strict criteria that RAPD data must satisfy if they were to be used to estimate population genetic parameters. If these criteria are not met then the frequency measure may be biased. Such criteria may also be applied to AFLP data, and it is unlikely that all or even a majority of these criteria would be met in an average AFLP study.
Character measures. Character measures would seem an ideal way of analysing AFLP data. AFLPs are easily scored as a discrete binary data matrix, the characters being the AFLP bands and their states either present or absent. The literature indicates that changing this sort of data into either similarity or frequency measures is undesirable (Swofford et al., 1997), primarily because character measures contain more "information" than distance measures.
Swofford and Olsen (1990) suggest that to be used as character data, characters must be variable, independent and homologous. AFLPs are variable, but some AFLP bands are non-homologous and non-independent. There is no way in which to identify and remove these non-homologous and non-independent bands from the data matrix a priori; both deficiencies will compromise an analysis.
The AFLP data, if used as characters, may be analysed using parsimony or maximum likelihood methods. The use of maximum likelihood methods for the analysis of AFLP data is not at present possible. The maximum likelihood (ML) method would model the processes that cause the gain or loss of AFLP bands and assign likelihoods or probabilities to these events. A tree would then be constructed that was the most likely, as the ML method does for sequence analysis, where the processes involved with nucleotide change can be modelled accurately (Page and Holmes, 1998). Such investigations have not taken place for AFLP data.
Backeljau et al. (1995) discussed the choice of parsimony model for RAPD analyses, but many of the points are applicable to AFLP analyses. Few analyses of AFLP data have used parsimony, e.g. Kardolus et al. (1998), investigating the systematics of Solanum (Solanaceae), implemented Wagner parsimony, whilst Angiolitto et al. (1999) investigating genetic diversity in Olea (Oleaceae) implemented Dollo parsimony. Swofford et al. (1997) reviewed the different types of parsimony. Wagner parsimony, one of the simplest, assumes free reversibility of characters, which with just two characters means that the loss and gain of AFLP band are assumed to occur at identical rates. However, the two events may not happen with the same probability. An alternative is Dollo parsimony, which allows reversals (band loss), but will allow gains to occur only once on a tree. Again this is unrealistic, as the probability of an independent gain of a band is not negligible. The unreality of both Wagner and Dollo parsimony to the biological data has been considered for RFLP analysis (Wendel and Albert, 1992). In the case of RFLPs, weighted parsimony has been implemented (Wendel and Albert, 1992). In this form of parsimony, the different probabilities of the gain or loss of a site are assigned. With RFLPs, and especially plastid RFLPs, the processes causing the gain/loss of a restriction site are fairly well understood and this has allowed workers to assign weight to the gain or loss of a site, e.g. the gain of a site restriction enzyme site is weighted twice as heavily as the loss of a site (Wendel and Albert, 1992).
Although the processes involved in the gain or loss of a AFLP may appear similar to those creating RFLPs, there are several features in which these differ. RFLP analyses that use the site occurrence method of Bremer (1991) are able to remove non-homologous and non-independent fragments from the matrix (or analyse them separately). The AFLP method amplifies fragments from throughout the genome [similar to the fragment occurrence analysis of Bremer (1991)] making it impossible to "type" the fragments., which leaves non-homologous and non-independent characters in the matrix. Bremer (1991) found that fragment methods were less accurate than other methods. In addition, the nature of the amplification, although much more reliable than the RAPD reaction, is not well understood, with different patterns found in re-amplifications and "template independent bands" (Vos et al., 1995) amongst these factors. Until there is a better understanding of the AFLP reaction it seems inappropriate to consider AFLP products as character data in parsimony analyses.
Distance Measures and Phylogenetic Reconstruction.
Objections may be raised to the use of AFLP data as character data, and the inapplicability of parsimony methods of phylogenetic reconstruction in these cases. However, more frequent is the use of similarity measures to cluster accessions together from which phylogenetic hypotheses are proposed. For example, Sharma et al. (1996) used a Nei-Li estimate of similarity and the neighbourhood-joining method to produce a dendrogram, and their results "supported" the classifications of Lens based on other data.
The use and applicability of cluster dendrograms to inference of phylogenies is a debate that started in the Sixties with the introduction of phenetics and the emergence of cladistics and continues to this day, although Page and Holmes (1998) point out that just because a method is phenetic does not mean it is meaningless.
The methods used to construct dendrograms from distance matrices are well understood (e.g. Avise, 1995; Swofford et al. 1997). Using distance measures to construct a dendrogram from AFLP data consists of two steps: i) the conversion of the binary data matrix to a distance matrix; and ii) the use of the distance matrix and a tree building programme to construct the dendrogram. Tree building programmes are simple algorithms and will build a dendrogram regardless of the data quality. The use of AFLP distance matrices and phylogenetics is centred on the quality of the data entering the tree-building programmes. As discussed above there are many possible causes of error in an AFLP analysis, and these may cause problems with the calculation of genetic distances that adds a level of uncertainty to the resulting dendrogram.
Microsatellites.
Microsatellites, alternatively known as simple sequence repeats (SSRs), short tandem repeats (STRs) or simple sequence length polymorphisms (SSLPs), are tandem repeats of sequence units generally less than 5 bp in length, e.g. (TG)n or (AAT)n (Bruford and Wayne, 1993). These markers appear to be hypervariable, in addition to which their co-dominance and reproducibility make them ideal for genome mapping, as well as for population genetic studies (Dayanandan et al., 1998). Inter-SSRs are a variant of the RAPD technique, although the higher annealing temperatures probably means that they are more rigorous than RAPDs. Chloroplast microsatellites (cpSSRs), are similar to nuclear microsatellites but the repeat is usually only 1 bp, i.e. (T)n (e.g. Provan et al., 1999).
What are Microsatellites and how are they produced?
Microsatellite variation results from differences in the number of repeat units. These differences are thought to be caused by errors in DNA replication (Moxon and Willis, 1999; Jarne and Lagoda, 1996); the DNA polymerase "slips" when copying the repeat region, changing the number of repeats (Jarne and Lagoda, 1996). Larger changes in repeat number are though to be the result of processes such as unequal crossing over (Strand et al., 1993). Such differences are detected on polyacrylamide gels, where repeat lengths migrate different distances according to their sizes.
Methods for screening and detecting microsatellites are covered extensively elsewhere (Ciofi et al., 1998; Rafalski et al., 1996; Powell et al, 1995; White and Powell, 1997b; Connell et al., 1998; Ciaferelli et al. 1995; Lench et al., 1996). The microsatellite protocol is simple, once primers for SSRs have been designed. The first stage is a PCR, depending upon the method of detection one of the primers is fluorescently or radioactively labelled. The PCR products are separated on a high resolution polyacrylamide gels, and the products detected with a fluorescence detector (e.g. automated sequencer) or an X-ray film.
How have microsatellites been used?
Microsatellites, which detect variation at individual loci, have been thought of as the "new allozymes". Consequently much of their use has been in studies where allozymes have been used, e.g. diversity studies (e.g. Rosette et al., 1999), gene-flow and mating (Chase et al., 1996) systems and paternity analysis (Streiff et al., 1999). Rosette et al. (1999) studied the partitioning of variation within and between populations of Melaleuca alternifolia (Myrtaceae) to facilitate the identification of genetic resources and assist in the conservation of genetic diversity. Chase et al. (1996) studied the gene-flow and mating patterns of Pithecellobium elegans (Leguminosae) in a forest fragment in Costa Rica, whilst Aldrich et al. (1998) analysed the genetic structure and diversity of fragmented populations of Symphonia globulifera (Clusiaceae). However, there are few phylogenetic studies that use microsatellite markers.
Many microsatellite studies appear to be expansions of groups that have been studied using biochemical or molecular markers. Rossetto et al.'s (1999) study on genetic structure in Melaleuca alternifolia is an expansion of Butcher et al.'s (1992) allozyme studies, albeit Rossetto et al. (1999) used a greater number of individuals and populations. Other studies have taken advantage of the high variability of microsatellites to re-study species in which previous methods have found little or no variation. For example, little variation has been found in Pinus resinosa using allozymes or RAPDs, but a study using chloroplast microsatellites (cpSSRs, Echt et al., 1998), found 23 different haplotypes using nine cpSSR loci.
Other studies have assessed cross-species amplification of microsatellite primers. Many initial microsatellite studies have been confined to single taxa; the taxon for which the primers were developed. The reason for this appears to be due to the perceived inability of microsatellite primers to amplify DNA in species other than the one in which they were "typed"; which may be why few systematic studies using microsatellites have been published. However, some studies have indicated that SSR primers may amplify the same SSR region in closely related taxa. For example, White and Powell (1997a) surveyed the Meliaceae, using primers designed for Swietenia humulis (White and Powell, 1997b). They were able to amplify DNA from seven of the 11 microsatellite loci in other Swietenia species, six loci in other genera of the same tribe, and four to six loci in species of the same family. Other examples are Steinkellner et al. (1997) who described the conservation of microsatellite loci between Quercus species (Fagaceae) and Dayanandan et al. (1997) who investigated the conservation of loci in the tribe Ingeae (Leguminosae).
Such cross-species surveys show that it is possible to amplify SSRs from species other than those from used in the primer design. The extent of the cross-species amplification appears to be correlated with taxonomic distance, and the knowledge that some loci amplify across species has stimulated some phylogenetic studies. For example, Mhameed et al. (1997) studied the relationships between avocado (Persea americana, Lauraceae) and wild Persea species, and presented a phylogenetic tree derived using parsimony. Proven et al. (1999) used cpSSRs in a systematic and population genetic study of the genus Hordeum (Poaceae), using a phenetic measure to construct a phylogenetic tree.
Advantages.
As with AFLPs, the great advantage of microsatellite analysis is the large number of polymorphisms that the method reveals. One locus in soybean (Glycine max) is reported to have 26 alleles (Cregan et al., 1994). Furthermore, the ability of the method to differentiate individuals when a combination of loci is examined makes the technique very useful for gene-flow experiments, cultivar identification and paternity analyses (Hokanson et al., 1998). Since microsatellites only survey one loci at a time, they are not directly comparable to AFLPs, for example Maughan et al. (1998) found that AFLPs produced more polymorphic loci than SSRs. Comparisons that include microsatellites should be with other single loci markers, such as RFLPs and isozymes. For example, Rossetto et al. (1999) found observed heterozygosity (Ho) for Melaleuca alternifolia microsatellites to be 0.724, much higher than the value for allozymes (Ho = 0.154; Butcher et al., 1992). McCouch et al. (1997) compared the number of alleles revealed by RFLPs and microsatellite loci in rice (Oryza spp.), and found 2-25 alleles per microsatellite loci compared with 2-4 alleles per RFLP loci, illustrating the large number of polymorphisms potentially highlighted by microsatellites.
Unlike AFLPs, microsatellites are co-dominant markers, thus heterozygotes can be readily identified. Microsatellite co-dominance will increase the efficiency and accuracy of population genetic measures based on these markers compared with other markers, such as AFLPs and RAPDs. Furthermore, the identity of heterozygotes in the F1 generation makes gene-flow, hybridisation and paternity analyses simpler (Schlötterer and Pemberton, 1994).
Since the method is DNA-based, this brings advantages, such as high-throughput and the ability to use dried leaf material. In comparison with allozymes, SSRs are thought to be selectively neutral, which though not essential for phylogenetic studies is one of the assumptions of using markers in many analyses.
Once SSR primers have been identified, screening of material using the technique is fairly inexpensive. Furthermore, cross-species amplification of SSRs means that identification of suitable SSR primers may not be necessary in closely related taxa. For example, three sets of microsatellite primers have been designed in Malus domestica (Rosaceae), which yield 35 loci, some of which may amplify other Malus taxa (Guilford et al., 1997; Gianfranceschi et al., 1998; Hokanson et al., 1998).
Problems.
As with the problems associated with AFLPs, those relating to microsatellites may be divided into three broad categories.
Practical.
Screening for SSRs. Unless useful primers have been designed in previous studies, it is necessary to screen an organism for microsatellites. There are many different ways of screening, all of them are practically complex and expensive and may yield only a small number of potential microsatellite loci. For example, Kelley and Willis (1998) screened 150,000 plaques with a SSR probe, and of these 179 positive plaques were sequenced.
"Slippage". This can be a significant problem when analysing mono- and di-nucleotide repeats (Ciofi et al., 1998). During the amplification process the thermopolymerase can "slip", leading to the production of differently sized products (Ciofi et al., 1998) that differ by approximately 1-5 repeat units from the expected product. Such products are usually less intense than the desired product, so in practice can usually be discounted. However, if the products of a heterozygous individual overlap then it is sometimes difficult to differentiate the true and slippage products (Ciofi et al., 1998).
Additional practical problems. Haberl & Tautz (1999) and Chavarriaga et al. (1998) both highlight a potential problem with SSRs run on automatic sequencing gels and automatically sized. Haberl & Tautz (1999) found that the "called" product sizes differed from the exact product sizes, for example, a real difference between alleles of 18 nucleotides was "called" by the computer as a mean of 16.68 nucleotides. Between AFLP gels, Chavarriaga et al. (1998) found a mean difference of 1.04 nucleotides, with a maximum "called" difference of 2.17 nucleotides. Haberl & Tautz (1999) recommended that exact sizes could only be determined by allele sequencing and determining "real size" and then using these as internal standards on a gel.
Inaccurate allele identification may also be caused by the tendency of Taq polymerase to add an adenosine nucleotide to the 3'-end of the amplified product (Ciofi et al., 1998). The addition is determined in a template- and marker-specific manner, which may not be a problem if the extra nucleotide is always, or never, added. However, errors may occur in size determination if the extra nucleotide is only occasionally added, Smith et al. (1995) and Ginot et al. (1996) suggest ways of overcoming this problem.
Data Problems.
Homology. This is the greatest problem facing the use of SSRs in phylogenetic analyses. Microsatellite analyses assume that co-migrating fragments are homologous, whereas there are few a priori reasons to assume this. Furthermore, non-homology can be divided into that which occurs within the SSR flanking and the SSR repeat regions.
Several studies have sequenced amplified microsatellites to test homology and the mechanisms of microsatellite mutation (Blanquer-Maumont and Crouau-Roy, 1995; Grimaldi and Crouau-Roy, 1997; Buteler et al., 1999). Blanquer-Maumont and Crouau-Roy (1995) sequenced the microsatellites they amplified from humans and found variation in the non-repeated flanking regions. The variation consisted of both point mutations and indels. Point mutations will not change the length of a microsatellite product, but indels will change the length of an amplified product, possibly causing the length of the repeat to be misinterpreted. For example, Grimaldi and Crouau-Roy (1997) sequenced all the alleles from a (CA)n repeat from humans and found both point mutations and indels in the amplified fragment, e.g. they found a 6 bp insertion in the flanking region of the SSR. This mutation could possibly cause a misidentification of the alleles if only the size of the allele was measured, e.g. a (CA)20 allele containing the 6 bp insertion in its flanking region would co-migrate with a (CA)23 allele that did not have the insertion (Grimaldi and Crouau-Roy, 1997).
Buteler et al. (1999) characterised microsatellites in diploid and polyploid sweet potatoes (Ipomoea trifida and I. batatas, Convolvulaceae), and found "instability" in the microsatellite flanking regions. The "instability" in the non-repeated flanking regions consisted of both point mutations and indels, and occurred at a higher rate than in humans (Buteler et al., 1999; Callen et al., 1993). Buteler et al. (1999) suggested that caution should be used when relying exclusively on band size in the interpretation of SSR length polymorphisms. Ujino et al. (1998) also detected indels in the flanking regions of Shorea species. (Dipterocarpaceae). However, this contrasts with Steinkellner et al. (1997), who in a survey of SSR conservation in Quercus spp., amplified and sequenced 12 microsatellite bands (they do not say which loci the bands were from). Steinkellner et al. (1997) found all twelve bands to be "truly homologous" to the original microsatellite and flanking sequences.
Since no large-scale tests of SSR homology have taken place in plants, it is difficult to estimate the percentage of bands in a microsatellite survey that are non-homologous. However, on the basis of the above examples, there is the potential for a serious problem, and the inclusion of non-homologous fragments in an analysis is likely to bias the results and break the assumptions of a phylogenetic analysis.
Ujino et al. (1998) point out another homology problem that occurs when analysing compound repeats. If a SSR with the sequence 5'-(CT)10CA(CT)8-3' is considered and a second allele is 2 bp longer, without sequencing, it is impossible to tell which repeat has increased in size, i.e. 5'-(CT)11CA(CT)8-3' or 5'-(CT)10CA(CT)9-3'. One would expect a greater percentage of fragments to be non-homologous if the repeat being analysed were compound. This was recognised, when Ujino et al. (1998) recommended that only simple repeats be used in order to limit errors in genotype identification.
The third and most problematic homology uncertainty is within the repeat unit. That is, whether two fragments that co-migrate are identical by descent or just identical in state. There is no simple answer to this issue, and this does not seem to have been generally considered. Assuming that the stepwise mutation model (SMM) is a good way of describing the evolution of the SSRs, consider the case of two (CA)6 alleles. The (CA)6 allele will arise from either from a (CA)7 allele or (CA)5 allele. All accessions with a (CA)6 allele are assumed to be identical by descent, but there are few reasons to assume this. Six possible mutations will increase a (CA)5 allele to a (CA)6 allele (see Figure 2), plus an additional seven "mutations" that would decrease a (CA)7 allele to a (CA)6 allele.
The problem of homology depends upon the mutation rate of the repeats. If it is low then the probability that a mutation is unique and similar alleles are identical by descent is high. Vice versa, if mutation rate is high then the probability increases that two co-migrating alleles are just identical in state and non-homologous. As far as we can tell this value has not been estimated in plants, but reported values for mice and humans are of the order of 10-5-10-2 (Jarne and Lagoda, 1996), but if these high values are repeated in plants then the likelihood of non-homology from co-migrating alleles is high.
Doyle et al. (1998) measured the homoplasy of chloroplast microsatellites by mapping SSR alleles onto a phylogeny of Glycine derived from an extensive chloroplast restriction site survey. Doyle et al. (1998) contended that because of homoplasy, sizes (alleles) at microsatellite loci were poor markers for the haplotypes identified by Doyle et al. (1990). CpSSR alleles were not correlated with the putative phylogenetic position of the accessions surveyed, and Doyle et al. (1998) concluded that the microsatellite loci did not faithfully represent relationships among the genomes of Glycine.
Systematists have discussed the issue of paralogy (identical in state) versus orthology (identical by descent) for long time. Moritz and Hillis (1997) comment that the confusion of paralogous and orthologous sequences can result in a "a correctly estimated phylogeny for the molecules that differs markedly from that of the organisms from which they were sampled", and it is likely that the comparison of paralogous SSR alleles will suffer from the same problem, i.e. an incorrect organismal phylogeny.
The problem with the use of microsatellites in systematics is likely to be the large number of non-homologous, co-migrating alleles. Currently, estimates of the percentage of non-homologous alleles have not been made; flanking regions and compound repeats can be tested for non-homology by sequencing, but within microsatellite homology can only be assessed theoretically. The incidence of non-homology can bias similarity, frequency measures and character based measures (Swofford et al., 1997). As the number and percentage of homoplasious characters increases, so does the likelihood of error in the resultant phenetic and phylogenetic trees.
Null Alleles. Mutations in the binding region of one or both of the microsatellite primers may inhibit annealing that may result in the reduction or loss of the PCR product (Callen et al. 1993). Such products are termed null alleles and are comparable to the null alleles identified by allozymes in their effects.
Null alleles may be manifested as fewer heterozygotes than expected in a randomly mating population or by the appearance of "empty" lanes (Morgante et al., 1998). That is, in a heterozygote of two different microsatellite alleles, if one of these alleles cannot be amplified due to primer annealing difficulties, then the phenotype (on the SSR gel) will appear as a single banded homozygote. Null alleles are also responsible for mismatches between parent-offspring pairs, i.e. the offspring do not amplify an allele that is present in the parents (Pemberton et al., 1998). For example, Rossetto et al. (1999) found that for Melaleuca alternifolia, observed heterozygosity was lower than expected and a significant excess of homozygotes was found, which Rossetto et al. (1999) suggested is the result of null alleles, the Wahlund effect or partial selfing. Callen et al. (1993) found, that in a survey of (AC)n microsatellite markers in humans, that 7 (30%) of the 23 markers surveyed demonstrated the presence of null alleles. Callen et al. (1993) were able to detect null alleles through the non-inheritance, by a sib, of a parental allele. In plants the number of null alleles could be determined through the analysis of progeny arrays.
Rossetto et al. (1999) argued that since the primers they used were based on homologous primers in Melaleuca alternifolia, null alleles were less likely. However, Callen et al. (1993) identified null alleles using homologous primers. The use of heterologous primers is likely to increase the incidence of null allele detection. Direct evidence of this is plants is incomplete, but Simonsen et al. (1998) in a study of the African Buffalo (Syncerus caffer) using cattle primers (Bos taurus) found that three loci (from six sampled) significantly differentiated from Hardy-Weinberg equilibrium, due to an excess of homozygotes, this was explained by Simonsen et al. (1998) as due to a combination of null alleles and heterologous primers. When primers were redesigned for these loci to make them Buffalo-specific, two out of the three loci were in Hardy-Weinberg equilibrium. However, the frequency of null alleles in cross-species studies in plants is unknown.
Dowling et al. (1997) suggest that null alleles will bias the estimation of genotype and allele frequencies, whilst Lamboy (1994a) has shown that they will bias the estimation of distance or similarity measures. In cross-species studies it is likely that as the taxonomic distances between taxa increases then the incidence of null alleles will also increase. Clearly there is a need for empirical observation in this area.
Analysis problems.
Microsatellite variation may be analysed phylogenetically in two ways: i) presence or absence of alleles as characters, and calculating either distance or using character measures; and ii) allele frequency at loci as characters and calculating distance measures. The coding of allozymes for phylogenetic analyses is very similar, and much of this extensive discussion is relevant to microsatellites (Buth, 1984; Murphy, 1993; Swofford et al., 1997).
Presence/Absence. Murphy (1993) argued that the presence/absence method was an invalid means of analysing data since: i) independent losses of "primitive" alleles are considered as synapomorphies; ii) loci with a greater number of alleles are given a greater weight in tree reconstruction; iii) unnecessary character conflicts arise when no alleles are shared between the ingroup and outgroup; and iv) outgroup polymorphism may result in erroneous hypotheses. Presence/absence data may be converted into either a pair-wise similarity matrix (e.g. Provon et al., 1999) or analysed as character data (e.g. Mhameed et al., 1997). Murphy's (1993) objections will result in bias when either method is used for estimating relationships, and in the case of parsimony methods there is the possibility that less parsimonious relationships may be found. SSRs introduce additional sources of error, through the potential non-homology of the SSR bands and the presence of null alleles.
Allele frequency. The calculation of allele frequencies at each locus has not been extensively used for phylogenetic analyses, partly because of the difficulties in data coding and computing distances (Buth and Murphy, 1999). Part of the discussion of frequency measures is taken up with a discussion of whether the model of repeat evolution should be the infinite allele or stepwise mutation model (Jarne and Lagoda, 1996). As might be expected the SMM appears to be consistent with the observed allele frequencies at SSR loci (Valdes et al., 1993). Di Rienzo et al. (1994) suggested a two-phase model, in which the primary changes are single addition or losses of repeats with the occasional rare large change in repeat number. Swofford and Berlocher (1987) describe a method of inferring trees, and Berlocher and Swofford (1997) discuss the practicalities of using such a method. Farris (1981, 1985) argues against the use of allele data for phylogenetic reconstruction, but his arguments are based primarily on the inappropriateness of the analyses. Crother (1990) suggests that it is the "nature" of allele frequencies which should prevent their use in phylogenetic analyses, and focuses on the opinion that the allele frequencies are not temporally stable, and so therefore cannot be synapomorphic; furthermore, the effects of non-homologous and null alleles on the derivation of phylogenies unknown.
Conclusions.
No single objection to the use of AFLPs for systematic studies exists, but the weight of circumstantial evidence cautions against their use. Firstly, the problems of non-homology and non-independence of the AFLP data have the potential to seriously mis-estimate similarity and distance, and these two problems cannot be overcome without extensive testing. To which may be added the problems of scoring, bias introduced by dominance, reproducibility problems, the effect of polyploids, as well as practical problems. There are other methods of inferring phylogenies which are more rigorous, such as sequencing and plastid RFLPs, and which are no more expensive than AFLPs.
The use of SSRs for phylogenetic analyses is also unwarranted. The problem of the potential non-homology of co-migrating alleles, the presence of null alleles and the lack of any rigorous method of analysing the resultant data preclude their use in systematic studies. Jarne and Lagoda (1996) conclude their review of SSRs with the comment that microsatellites make very poor markers for phylogenetic inferences, except for "groups separated by no more than a few thousand generations".
For any problem under investigation it is the nature of the problem
that should dictate the method of analysis, the most modern method may
not always be the best or most cost-effective way of addressing the problem.
What is lacking, and is perhaps bringing about the use of AFLPs and SSRs
is a marker that can be used consistently below the species level for phylogenetic
studies; Schaal et al. (1998) discuss this and consider possible
solutions. We conclude that neither AFLPs nor microsatellites should be
considered for phylogenetic analyses above the species level. AFLPs and
SSRs are valuable methods for addressing population genetics and plant
breeding issues, but for phylogeny reconstruction and taxonomy they are
at best problematic and at worst misleading.
References
Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999) Phylogenetic relationships among Oryza species revealed by AFLP markers. Theoretical and Applied Genetics 98: 1320-1328.
Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Molecular Ecology 7: 933-944.
Angiolillo A, Mencuccini M, Baldoni L (1999) Olive genetic diversity assessed using amplified fragment polymorphisms. Theoretical and Applied Genetics 98: 411-421.
Avise JC (1994) Molecular Markers, Natural History and Evolution. Chapman and Hall, New York.
Backeljau T, de Bruyn L, de Wolf H, Jordaens K, Van Dongan S, Verhagen R, Winnepenninckx B (1995) Random amplified polymorphic DNA (RAPD) and parsimony methods. Cladistics 11: 119-130.
Barker JHA, Matthes M, Arnold GM, Edwards KJ, Åhman I, Larsson S, Karp A (1999) Characterisation of genetic diversity in potential biomass willows (Salix spp.) by RAPD and AFLP analyses. Genome 42: 173-183.
Beismann H, Barker JHA, Karp A, Speck T (1997) AFLP analysis sheds light on distribution of two Salix species and their hybrid along a natural gradient. Molecular Ecology 6: 989-993.
Berlocher SH, Swofford DL (1997) Searching for phylogenetic trees under frequency parsimony criteria: an approximation using generalised parsimony. Systematic Biology 46: 211-215.
Blanquer-Maumont A, Crouau-Roy B (1995) Polymorphism, monomorphism and sequences in conserved microsatellites in primate species. Journal of Molecular Evolution 41: 492-497.
Bradshaw JE, Hackett CA, Meyer RC, Milbourne D, McNichol JW, Philips MS, Waugh R (1998) Identification of AFLP and SSR marker associated with quantitative resistance to Globodera pallida (Stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theoretical and Applied Genetics 97: 202-210.
Bremer B (1991) Restriction data from cpDNA for phylogenetic reconstruction: is there only one accurate way of scoring? Plant Systematics and Evolution 175: 39-54.
Bruford MW, Wayne RK (1993) Microsatellites and their application to population genetic studies. Current Opinion in Genetics and Development 3: 939-943.
Butcher PA, Bell CJ, Moran GF (1992) Patterns of genetic diversity and nature of breeding system in Melaleuca alternifolia (Myrtaceae). Australian Journal of Botany 40: 365-375.
Buteler MI, Jarret RL, LaBonte DR (1999) Sequence characterisation of microsatellites in diploid and polyploid Ipomoea. Theoretical and Applied Genetics 99: 123-132.
Buth DG (1984) The application of electrophorectic data in systematic studies. Annual Review of Ecology and Systematics 15: 501-522.
Buth DG, Murphy RW (1999) The use of isozyme characters in systematic studies. Biochemical Systematics and Ecology 27: 117-129.
Callen DF, Thompson AD, Shen Y, Phillips HA, Richards RI, Mulley JC, Sutherland GR (1993) Incidence and origin of "Null" alleles in the (AC)n microsatellite markers. American Journal of Human Genetics 52: 922-927.
Castiglioni P, Ajmone-Marsan P, van Wijk R, Motto M (1999) AFLP markers in a molecular linkage map of maize: co-dominant scoring and linkage group distribution. Theoretical and Applied Genetics 99: 425-431.
Chase MR, Moller C, Kesseli R, Bawa KS (1996) Distant gene flow in tropical trees. Nature 383: 398-399.
Chavarriaga-Aguirre P, Maya MM, Bonierbale MW, Kresovich S, Fregene MA, Tohme J, Kochert G (1998) Microsatellites in Cassava (Manihot esculenta Crantz): discovery, inheritance and variability. Theoretical and Applied Genetics 97: 493-501.
Ciaferelli RA, Gallitelli M, Cellini F (1995) Random amplified hybridisation microsatellites (RAHM): isolation of a new class of microsatellite-containing DNA clones. Nucleic Acid Research 23: 3802-3803.
Ciofi C, Funk SM, Coote T, Cheesman DJ, Hammond RL, Saccheri IJ, Bruford MW (1998) Genotyping with microsatellite markers. In: Karp A, Isaac PG, Ingram DS (eds.). Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp. 195-201.
Clark AG, Lanigan CMS (1993) Prospects for estimating nucleotide divergence with RAPDs. Molecular Biology and Evolution 10: 1096-1111.
Connell JP, Pammi S, Iqbal MJ, Huizinga T, Reddy AS (1998) A high throughput procedure for capturing microsatellites from complex plant genomes. Plant Molecular Biology Reporter 16: 341-349.
Cregan PB, Bhagwat AA, Akkaya MS, Rongwen J (1995) Microsatellite fingerprinting and mapping in soybean. Methods in Molecular Cell Biology 5: 49-61.
Crother BI (1990) Is "some better than none" or do allele frequencies contain phylogenetically useful information? Cladistics 6: 277-281.
Dayanandan S, Rajora OP, Bawa KS (1998) Isolation and characterisation of microsatellites in trembling aspen (Populus tremuloides). Theoretical and Applied Genetics 96: 950-956.
Dayanandan S, Bawa KS, Kesseli R (1997) Conservation of microsatellites among tropical trees (Leguminosae). American Journal of Botany 84: 1658-1663.
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation, Version II. Plant Molecular Biology Reporter 1: 19-21.
DeVerno LL, Mosseler A (1997) Genetic variation in red pine (Pinus resinosa) revealed by RAPD & RAPD-RFLP analysis. Canadian Journal of Forest Research 27: 1336-1320.
DeBry RW, Slade NA (1985) Cladistic analysis of restriction endonuclease cleavage maps within a maximum likelihood framework. Systematic Zoology 34: 21-34.
Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proceedings of the National Academy of Science USA 91: 3166-3170.
Donini P, Elias ML, Bougourd SM, Koebner RMD (1997) AFLP fingerprinting reveals pattern differences between template DNA extracted from different plant organs. Genome 40: 521-526.
Dowling TE, Moritz C, Palmer JD, Rieseberg LH (1997) Nucleic Acids III: Analysis of fragments and restriction sites. In: Hillis DM, Moritz C, Mable BK (eds.). Molecular Systematics, 2nd. edition. Massachusetts, Sinauer Assoc. Inc., pp. 249-320.
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11-15.
Doyle JJ, Doyle JL, Brown AHD (1990) Chloroplast DNA polymorphism and phylogeny in the B genome of Glycine subgenus Glycine (Leguminosae). American Journal of Botany 77: 772-782.
Doyle JJ, Morgante M, Tingley SV, Powell W (1998) Size homoplasy in chloroplast microsatellites of wild perennial relatives of Soybean (Glycine subgenus Glycine). Molecular Biology and Evolution 15: 215-218.
Echt CS, DeVerno LL, Anzidei M, Vendramin GG (1998) Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Molecular Ecology 7: 307-316.
Escaravage N, Questiau S, Pornon A, Doche B, Taberlet P (1998) Clonal diversity in a Rhododendron ferrugineum L. (Ericaceae) population inferred from AFLP markers. Molecular Ecology 7: 975-982.
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferrred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479-491.
Farris, JS (1981) Distance data in phylogenetic analyses. In: Funk VA, Brooks DR (eds.). Advances in Cladistics: Proceedings of the first meeting of the Willi Henning Society. New York Botanical Garden, New York, pp. 2-23.
Farris JS (1985) Distance data revisited. Cladistics 1: 67-86.
Fowler DP, Morris RW (1977) Genetic diversity in red pine: evidence for low genetic heterozygosity. Canadian Journal of Forest Research 7: 343-347.
Francisco-Ortega J, Newbury HJ, Ford-Lloyd BV (1993) Numerical-analyses of RAPD data highlight the origin of cultivated tagasaste (Chamaecytisus proliferus ssp palmensis) in the Canary Islands. Theoretical and Applied Genetics 87: 264-270.
Gianfranceschi L, Seglias N, Tarchini R, Komjanc M, Gessler C (1998) Simple sequence repeats for the genetic analysis of apple. Theoretical and Applied Genetics 96: 1069-1076.
Gift N, Stevens PF (1997) Vagaries in the delimitation of character states in quantitative variation- an experimental study. Systematic Biology 46: 112-125.
Ginot F, Bordelais I, Nguyen S, Gyapay G (1996) Correction of some genotyping errors in automated fluorescent microsatellite analysis by enzymatic removal of one base overhangs. Nucleic Acids Research 24: 540-541.
Grimaldi M-C, Crouau-Roy B (1997) Microsatellite allelic homoplasy due to variable flanking sequences. Journal of Molecular Evolution 44: 336-340.
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus x domestica (apple): abundance polymorphism and cultivar identification. Theoretical and Applied Genetics 94: 249-254.
Haberl M, Tautz D (1999) Comparative allele sizing can produce inaccurate allele size differences for microsatellites. Molecular Ecology 8: 1347-1350.
Harris SA (1999a) RAPDs in systematics - a useful methodology? In: Hollingsworth PM, Bateman RM, Gornall RJ (eds.). Molecular Systematics, Plant and Evolution. Taylor and Francis, London, pp. 221-228.
Harris SA (1999b) Molecular approaches to assessing plant diversity. In: Benson EE (ed.). Plant Conservation Biotechnology. Taylor and Francis, London, pp. 11-24.
Harris SA, Robinson J (1994) Preservation of Tropical Plant Material for Molecular Analyses. In: Adams (ed.). Conservation of Plant Genes II. Monographs. Syst. Bot. Missouri Bot. Gard. No. 48. St Louis, Missouri Botanical Garden, pp. 83-92.
Hartl L, Seefelder S (1998) Diversity of selected hop cultivars detected by fluorescent AFLPs. Theoretical and Applied Genetics 96: 112-116.
Heun M, Schäfer-Pregl R, Klawan D, Castagna R, Accerbi M, Borghi B, Salamini F (1997) Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278: 1312-1314.
Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus x domestica Borkh. core subset collection. Theoretical and Applied Genetics 97: 671-683.
Huang J, Sun M (1999) A modified AFLP with flourescence-labelled primers and automated DNA sequencer detection for efficient fingerprinting analysis in plants. Biotechnology Techniques 13: 277-278.
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bulletin de la Société Vaudense des Sciences Naturelles 44: 223-270.
Jarne P, Lagoda PJL (1996) Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution 11: 424-429.
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, van deWiel C, Bredemeijer G, Vosman D, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevshi A, Marmiroli N, Aert R, Volchaert G, Rueda J, Linacerro R, Vazquez A, Karp A (1997) Reproducibility testing of RAPD, AFLP and SR markers in plants by a network of European laboratories. Molecular Breeding 3: 381-390.
Kardolus JP, van Eck HJ, van den Berg RG (1998) The potential of AFLPs in biosystematics: a first application in Solanum taxonomy (Solanaceae). Plant Systematics and Evolution 210: 87-103.
Karp A, Kresovich S, Bhat KV, Ayand WG, Hodgkin T (1997) Molecular tools in plant genetic resources conservation: a guide to the technologies. IPGRI Technical Bulletin No. 2, International Plant Genetic Resources Institute, Rome, Italy. Available at http://198.93.227.125/publicat/techbull/TB2.pdf.
Kelley AJ, Willis JH (1998) Polymorphic microsatellite loci in Mimulus guttatus and related species. Molecular Ecology 7: 769-774.
Krauss SL, Peakall R (1998) An evaluation of the AFLP fingerprinting technique for the analysis of paternity in natural populations of Persoonia mollis (Proteaceae). Australian Journal of Botany 46: 533-546.
Krauss SL (1999) Complete exclusion of nonsires in an analysis of paternity in a natural plant population using amplified fragment length polymorphism (AFLP). Molecular Ecology 8: 217-226.
Lamboy W (1994) Computing genetic distance similarity coefficients from RAPD data: the effects of PCR artifacts. PCR Methods and Applications 4: 31-37.
Law JR, Donini P, Koebner RMD, Jones CR, Cooke RJ (1998) DNA profiling and plant variety registration III: The statistical assessment of distinctness in wheat using amplified fragment length polymorphisms. Euphytica 102: 335-342.
Lench NJ, Norris A, Bailey A, Booth A, Markham AF (1996) Vectorette PCR isolation of microsatellite repeat sequences using anchored dinucleotide repeat primers. Nucleic Acid Research 24: 2190-2191.
Lerceteau E, Szmidt AE (1999) Properties of AFLP markers in inheritance and genetic diversity studies of Pinus sylvestris L. Heredity 82: 252-260.
Lewis PO, Snow AA (1992) Deterministic paternity exclusion using RAPD markers. Molecular Ecology 1: 155-160.
Lin JJ, Kuo J (1995) AFLP, a novel PCR-based assay for plant and bacterial DNA fingerprinting. Focus 17: 66-70.
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Molecular Ecology 3: 91-99.
Mace ES, Gebhardt CG, Lester RN (1999) AFLP analysis of genetic relationships in the tribe Datureae (Solanaceae). Theoretical and Applied Genetics 99: 634-641
Matthes MC, Daly A, Edwards KJ (1998) Amplified fragment length polymorphism (AFLP). In: Karp A, Isaac PG, Ingram DS (eds.). Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp. 183-190.
Maughan PJ, Saghai Maroof MA, Buss GR (1996) Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theoretical and Applied Genetics 93: 392-401.
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Molecular Biology 35: 89-99.
Mhameed S, Sharon D, Kaufman D, Lahav E, Hillel J, Degani C, Lavi U (1997) Genetic relationships within avocado (Persea americana Mill.) cultivars and between Persea species. Theoretical and Applied Genetics 94: 279-286.
Morgante M, Pfeiffer A, Jurman I, Paglia G, Olivieri AM (1998) Isolation of microsatellite markers in plants. In: Karp A, Isaac PG, Ingram DS (eds.). Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp. 288-296.
Moritz C, Hillis DM (1997) Molecular Systematics: context and controversies. In: Hillis DM, Moritz C, Mable BK (eds.). Molecular Systematics, 2nd. edition. Massachusetts, Sinauer Assoc. Inc., pp. 1-13.
Moxon ER, Wills C (1999) DNA microsatellites: agents of evolution? Scientific American, January: 72-77.
Muluvi GM, Sprent JI, Soranzo N, Provan J, Odee D, Folkard G, McNicol JW, Powell W (1999) Amplified fragment length polymorphism (AFLP) analysis of genetic variation in Moringa oleifera Lam. Molecular Ecology 8: 463-470.
Murphy RW (1993) The phylogenetic analysis of allozyme data: invalidity of coding alleles by presence/absence and recommended procedures. Biochemical Systematics and Ecology 21: 25-38.
Nakajima Y, Oeda K, Yamamoto T (1998) Characterisation of genetic diversity of nuclear and mitochondrial genomes in Daucus varieties by RAPD and AFLP. Plant Cell Reports 17: 848-853.
Nei M, Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Science 76: 5269-5273.
Page RD, Holmes EC (1998) Molecular Evolution: A Phylogentic Approach. Blackwell Science Ltd., Oxford.
Pemberton JM, Slate J, Bancroft DR, Barrett JA (1995) Nonamplifying alleles at microsatellite loci: a caution for parentage and population studies. Molecular Ecology 4: 249-252.
Perera L, Russell JR, Provan J, McNicol JW, Powell W (1998) Evaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling. Theoretical and Applied Genetics 96: 545-550.
Powell W, Morgante M, McDevitt R, Vendramin G, Rafalski A (1995) Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proceedings of the National Academy of Science USA 92: 7759-7763.
Powell W, Morgante M, Andre C, Hanafey M, Vogel MJ, Tingey SV, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellites) markers for germplasm analysis. Molecular Breeding 2: 225-235.
Proven J, Russell JR, Booth A, Powell W (1999) Polymorphic chloroplast simple sequence repeat primers for systematic and population studies in the genus Hordeum. Molecular Ecology 8: 505-511.
Qi X, Lindhout P (1997) Development of AFLP markers in barley. Molecular and General Genetics 254: 330-336.
Rafalski JA, Vogel JM, Morgante M, Powell W, Andre C, Tingey SV (1996) Generating and using DNA markers in plants. In: Birren B, Lai E (eds.). Non-Mammalian Genomic Analysis: A Practical Guide. Academic Press, London. pp. 75-134.
Ridout CJ, Donini P (1999) Use of AFLP in cereals research. Trends in Plant Science 4: 76-79.
Rieseberg LH (1996) Homology among RAPD fragments in interspecific comparisons. Molecular Ecology 5: 99-103.
Rieseberg LH, Kim MJ, Seiler GJ (1999) Introgression between the cultivated sunflower and a sympatric wild relative, Helianthus petiolaris (Asteraceae). International Journal of Plant Science 160: 102-108.
Rossetto M, Slade RW, Baverstock PR, Henry RJ, Lee LS (1999) Microsatellite variation and assessment of genetic structure in teatree (Melaleuca alternifolia - Myrtaceae). Molecular Ecology 8: 633-643.
Rouppe van der Voort JNA, van Zandvoort P, van Eck HJ, Folkertsma RT, Hutten RCB, Draaistra J, Gommers FJ, Jacobsen E, Helder J, Bakker J (1997) Use of allele specificity of comigrating AFLP markers to align genetic maps from different potato genotypes. Molecular and General Genetics 255: 438-447.
Russell JR, Weber JC, Booth A, Powell W, Sotelo-Montes C, Dawson IK (1999) Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon Basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Molecular Ecology 8: 199-204.
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7: 465-474.
Schlötterer C, Pemberton J (1994) The use of microsatellites for genetic analysis of natural populations. In: Schierwater B, Streit B, Wagner GP, DeSalle R (eds.). Molecular Ecology and Evolution: Approaches and Applications, Birkhäuser Verlag Basel, Switzerland, pp. 203-214.
Sharma SK, Knox MR, Ellis THN (1996) AFLP analysis of the diversity and phylogeny of Lens and its comparison with RAPD analysis. Theoretical and Applied Genetics 93: 751-758.
Simon J-P, Bergeran Y, Gagnon D (1986) Isozyme uniformity in populations of red pine (Pinus resinosa) in the Abitibi Region, Quebec. Canadian Journal of Forest Research 16: 1133-1135.
Simonsen BT, Siegismund HR, Arctander P (1998) Population structure of African buffalo inferred from mtDNA sequences and microsatellite loci: high variation but low differentiation. Molecular Ecology 7: 225-237.
Smith JR, Carpten JD, Brownstein MJ, Ghosh S, Magnuson VL, Gilbert DA, Trent JM, Collins FS (1995) Approach to genotyping errors caused by nontemplated nucleotide addition by Taq DNA-polymerase. Genome Research 5: 312-317.
Sneath PHA, Sokal RR (1973) Numerical Taxonomy. Freeman, San Francisco.
Steinkellner H, Lexer C, Turestschek E, Glössl J (1997) Conservation of (GA)n microsatellite loci between Quercus sp. Molecular Ecology 6: 1189-1194.
Strand M, Prolla TA, Liskay RM, Petes TD (1993) Destabilisation of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274-276.
Streiff, R, Ducousso A, Lexer C, Steinkellner H, Glössl J, Kremer A (1999) Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Quercus petraea (Matt.) Liebl. Molecular Ecology 8: 831-841.
Struss D, Plieske J (1998) The use of microsatellite markers for detection of genetic diversity in barley populations. Theoretical and Applied Genetics 97: 308-315.
Stuessy TF (1990) Plant Taxonomy: the systematic evaluation of comparative data. Columbia University Press, New York.
Swofford DL, Olsen GJ (1990) Phylogeny Reconstruction. In: Hillis DM, Moritz C (eds.). Molecular Systematics, Sinauer Assoc. Inc, Massachusetts, pp. 407-514.
Swofford DL, Olsen GJ, Waddell PJ, Hillis DM (1997) Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK (eds.). Molecular Systematics, 2nd. edition. Sinauer Assoc. Inc., Massachusetts, pp. 407-514.
Swofford DL, Berlocher SH (1987) Inferring evolutionary trees from gene frequency data under the principle of maximum parsimony. Systematic Zoology 36: 293-325.
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105-1109.
Travis SE, Maschinski J, Keim P (1996) An analysis of genetic variation in Astragalus cremnophylax var. cremnophylax, a critically endangered plant, using AFLP markers. Molecular Ecology5: 735-745.
Ujino T, Kawahara T, Tsumura Y, Nagamitsu T, Yoshimaru H, Ratnam W (1998) Development and polymorphism of simple sequence repeat DNA markers for Shorea curtisii and other Dipterocarpaceae species. Heredity 81: 422-428.
Valdes AM, Slatkin M, Freimer NB (1993) Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics 133: 737-749.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407-4414.
Wendal JF, Albert VA (1992) Phylogenetics of the cotton genus (Gossypium), character state weighted parsimony analysis of chloroplast DNA restriction site data and its systematic and biogeographic implications. Systematic Botany 17: 115-143.
White G, Powell W (1997a) Cross-species amplification of SSR loci in the Meliaceae family. Molecular Ecology 6: 1195-1197.
White G, Powell W (1997b) Isolation and characterisation of microsatellite loci in Swietenia humulis (Meliaceae): an endangered tropical hardwood species. Molecular Ecology 6: 851-860.
Winfield MO, Arnold GM, Cooper F, Le Ray M, White J, Karp A, Edwards KJ (1998) A study of genetic diversity in Populus nigra subsp. betulifolia in the Upper Severn area of the UK using AFLP markers. Molecular Ecology 7: 3-10.
Xu ML, Melchinger AE, Xia XC, Lübberstedt T (1999) High-resolution mapping of loci conferring resistance to sugarcane mosaic virus in maize using RFLP, SSR and AFLP markers. Molecular and General Genetics 261: 574-581.
Zhu J, Gale MD, Quarrie S, Jackson MT, Bryan GJ (1998) AFLP markers for the study of rice biodiversity. Theoretical and Applied Genetics 96: 602-611.
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Molecular Ecology 8: 907-913.
© Institut für Forstgenetik und Forstpflanzenzüchtung, Universität Göttingen, 1999